"...requires lower DNA input (25 ng) than current methods and is optimized for lower quality samples" - Childhood Cancer Data Initiative: Status report published by National Cancer Institute 2024"
〰️
"...requires lower DNA input (25 ng) than current methods and is optimized for lower quality samples" - Childhood Cancer Data Initiative: Status report published by National Cancer Institute 2024" 〰️
FORSHEAR™>SRSLY for degraded genomic DNA
enzymatic shearing for short read sequencing
Quality of genomic DNA is highly dependent on the source, storage and extraction methods. DNA from formalin-fixed paraffin embedded (FFPE) samples and herbarium specimens can be highly degraded and damaged. These inputs types perform poorly with tradiitonal NGS library preparation methods.
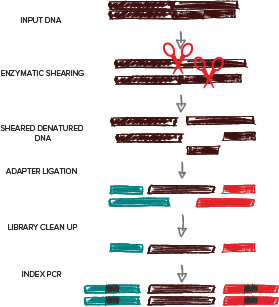
By combining our flagship library prep SRSLY with a robust enzymatic DNA shearing module ForShear, we now provide an optimized workflow that generates high-quality sequencing libraries from gDNA that are compatible with Illumina® sequencing platforms, forshear!
Forshear overview
Keeping to our theme of single-tube workflows, ForShear enzymatic fragmentation is performed in the same tube as single-strand stabilization steps with our ssEnhancer cocktail. Heat-inactivation of the ForShear enzymes also denatures the input DNA into single-strand - a prerequisite of SRSLY. This eliminates the need for bead purification or additional steps. And as SRSLY captures ssDNA, nicked DNA and native ends, this results in high library conversion metrics.
KIT COMPONENTS
-ForShear™ Shearing Enzyme
-ForShear™ Activity Buffer
-ForShear™ Dilution Buffer
Performance RANGE
10-50ng
DIN as low as 2
input types
FFPE DNA
gDNA
High molecular weight viral DNA
SRSLY with forshear for FFPE DNA
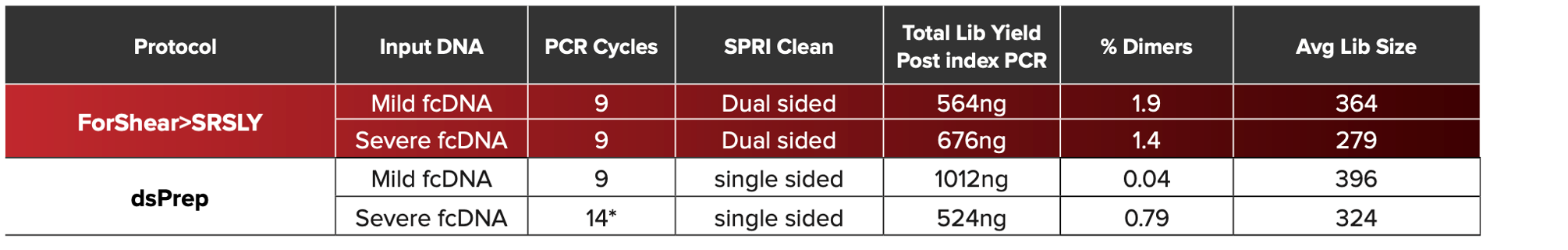
ForShear>SRSLY Molecular Metrics. Libraries were prepared with the Horizon Discovery formalin compromised DNA set - Mild fcDNA (DIN ~7) and Severe fcDNA (DIN ~3). For Mild fcDNA, both SRSLY>ForShear protocol and the dsPrep protocol showed equivalent molecular metrics. However, for the severe fcDNA, SRSLY>ForShear generated high-quality libraries at lower PCR cycles with minimal adapter dimers. Overall, the SRSLY approach generated smaller libraries presumably due to the capture of nicked and short ssDNA. Download the collateral for more information.
ForShear>SRSLY Whole Genome Sequencing Metrics. The mapping metrics of the libraries showed that both approaches generated good quality libraries for both input types with comparable chimeric rates, genome coverage and GC content. However, ForShear>SRSLY generated libraries with lower duplication rates and higher complexity even with the severely damaged input types.
ForShear>SRSLY Whole Exome Enrichment Metrics- Here performed exome-enrichment with the Twist® Exome 2.0 capture panel after library prep with ForShear>SRSLY NanoPLus with UMI Addition. Deconvolution was performed using the srslyumi pipeline. Overall, the ClaretBio workflow had lower mean error rate based on consensus UMI groups. For severe fcDNA, the dsPrep showed lower mean target coverage and lower observed concordance with the expected allele frequency based on ddPCR.
compatibility
DOWNSTREAM TARGETED ENRICHMENT
The Forshear + SRSLY workflow along with our UMI Additional module generates libraries of high complexity. This improves the accuracy of variant calling from degraded DNA of DIN as low as 2 from 10 ng inputs. Download the brochure to learn more. Write to us for sample data.
AUTOMATION FRIENDLY
As an enzymatic fragmentation method, Forshear does not require instrumentation and be run on a variety of liquid handler. Read this app note or visit our automation page
Want More Forshear?
Watch these videos to learn more about the Forshear>SRSLY workflow, applications and more.
In this webinar presentation, Dr. Katie Miller talks about the application of SRSLY with Forshear for limiting amounts of DNA from pediatric patients with focal epilepsy
Watch this SERIOUS TALK webinar by ClaretBio CSO Dr. Chris Troll to learn more about DNA shearing methods for NGS